Results
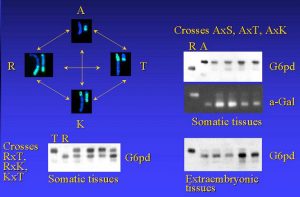
Nonrandom X-chromosome Inactivation in the F1 hybrids of common voles. A - M.arvalis; R (S) - M.rossiameridionalis; K - M.kirgisorum; T - M. transcaspicus
There is a phenomenon of the nonrandom X chromosome inactivation in some common voles (genus Microtus, “arvalis”) F1 hybrids, correlates with the presents of heterochromatin block on inactivated X-chromosome. Crosses between M.arvalis and another species of «arvalis» group has active X-cromosome from M.arvalis. Extraembrionic tissues of F1 hybrids has imprinting inactivation of male X-chromosome.

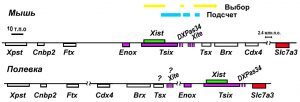
Mouse and vole Xist/Tsix locus. 1-8 – Xist exons; I-IV – mouse Tsix exones; A-D – M.arvalis Tsix;C’ – splising variant M.arvalis Tsix; pNif3l1 – pseudogene; DXPas34 –tandem repeat region; Xite – regulative mouse element.
We characterized the genomic Xist/Tsix locus for «arvalis» group species. Vole sequences downstream of the Xist gene show homology to minisatellite DXPas34 region and the major promoter of the Tsix gene of mouse Xic. We demonstrated that this putative promoter identified in vole is a site of origin of transcription. The antisence to the Xist transcription has practically the same pattern as the Tsix gene in mouse. High conservation of the Tsix major promoter sequences both in vole and mouse may be evidence of their crucial role in regulation of the Tsix gene transcription.
Commone voles Xic locus is reorganized compare to mouse Xic. In place of Xite gene (regulation of mouse Tsix expression, counting and choise element) there is Slc7a3 gene (2,5 m.b.p. out of mouse Xic centre) in vole Xic.
The gene Tsix minor promoter and the regulatory element Xite are deleted from X inactivation centre in vole as result of a chromosomal inversion. The fact allows suggesting that both the nucleotide sequences of the elements and antisence transcription associated with them are not absolutely necessary for the X inactivation process even in rodents.

Localization Xist RNA in G-negative bends of commone vole (M.rossiameridionalis) X chromosom. Xist RNA -green; MS4 (heterochromatin vole DNA repeat) red)
Inactivated X chromosome (Xi) undergoes various chromatin modifications, which affect the transcription of genes and maintain their inactive state. The untranslated nuclear RNA encoded by Xist gene is a component of Xi chromatin in eutherians. Xist RNA spreads from the XIC locus of the inactivated X chromosome, coats it up, repressing transcription of the genes in cis, and recruits chromatin modifying proteins. We show that Xist RNA exhibits a banded pattern on the inactive X and is excluded from regions of constitutive heterochromatin. The banding pattern suggests a preferential association with gene-rich, G-light regions.
The Xi epigenetics marks (triMeH3K27, ubiquitynilated uH2A) are revealed only in G-negative bands. G-positive bands, regions of constitutive heterochromatin of Xa and Xi contain triМеН3К9 and Hp1 protein.
We obtained new in vitro TS (Trophoectoderm Stem) and Xen (eXtrembrional ENtoderm) cell system for studing imprinting X inactivation.
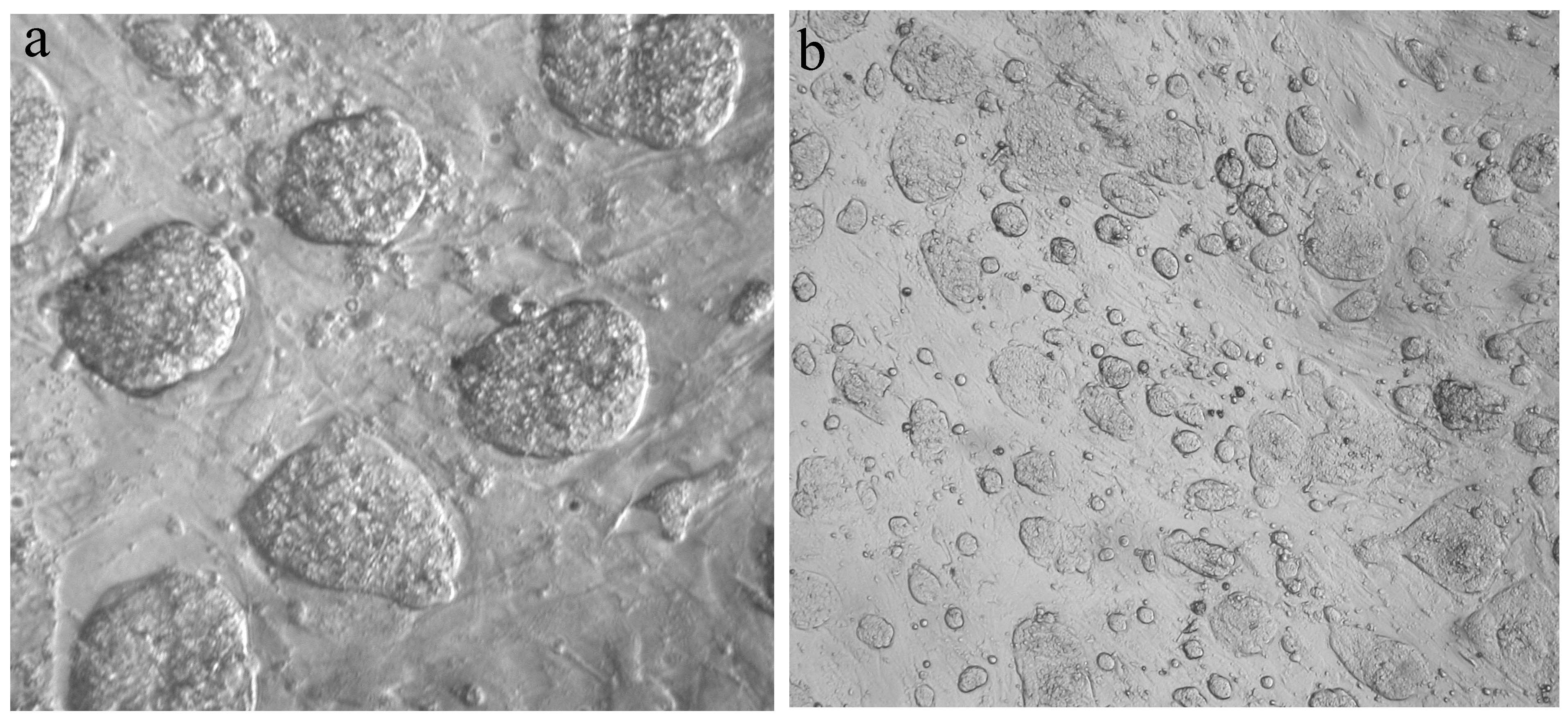
Common vole TS cell colonies on fider layer (primary mouse embrionic fibroblasts) a- 40x, b-8x magnification