Amyotrophic lateral sclerosis
Amyotrophic lateral sclerosis (ALS) is a neurodegenerative disease characterized by death of motor neurons. To date, neither etiology nor pathogenesis of ALS is clear, which leads to the absence of an effective treatment strategy. Animal models obtained through the years did not help to clarify the details of ALS initiation and propagation due to incomplete reproduction of the disease pathogenesis. Alongside with C9ORF72 hexanucleotide expansion and TDP-43 mutations, SOD1 mutations are the most common among patients with hereditary ALS. Notably, both clinical and molecular features of the disease are different between patients with mutations in different genes and even with different mutations in one gene. Since genetic background has a great impact on the pathology development it is important to keep it constant in order to study the effect of a particular mutation. To achieve this, we, using CRISPR/Cas9, have generated stem cell lines, containing Asp90Ala and Gly127Arg mutation in the SOD1 gene. These cell lines differ from each other and the original stem cell line only by the aforementioned mutations, so they can help to understand their impact on the ALS development.
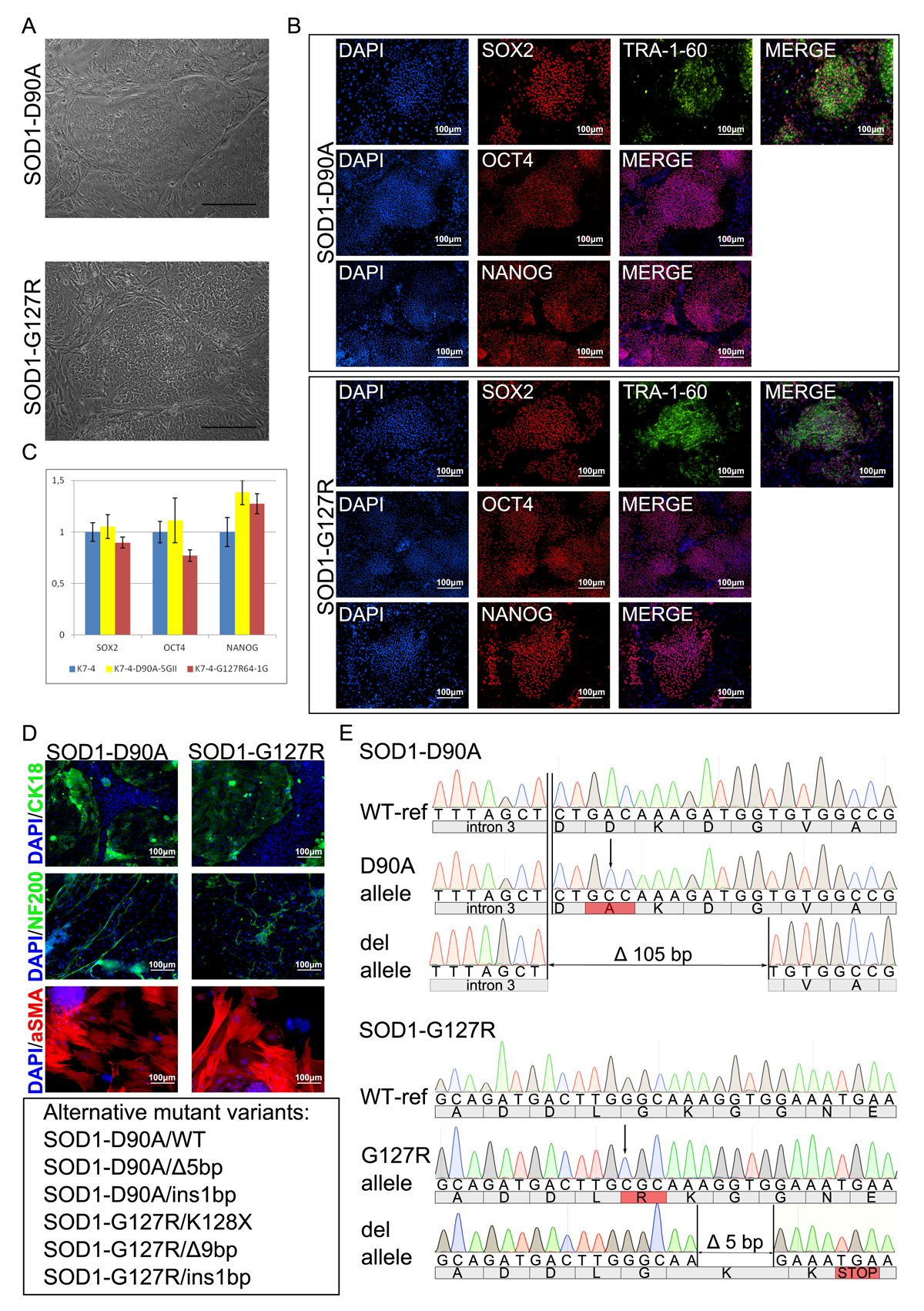
Amyotrophic lateral sclerosis (ALS) is a neurodegenerative disease characterized by death of motor neurons. To date, neither etiology nor pathogenesis of ALS is clear, which leads to the absence of an effective treatment strategy. Animal models obtained through the years did not help to clarify the details of ALS initiation and propagation due to incomplete reproduction of the disease pathogenesis. Alongside with C9ORF72 hexanucleotide expansion and TDP-43 mutations, SOD1 mutations are the most common among patients with hereditary ALS. Notably, both clinical and molecular features of the disease are different between patients with mutations in different genes and even with different mutations in one gene. Since genetic background has a great impact on the pathology development it is important to keep it constant in order to study the effect of a particular mutation. To achieve this, we, using CRISPR/Cas9, have generated stem cell lines, containing Asp90Ala and Gly127Arg mutation in the SOD1 gene. These cell lines differ from each other and the original stem cell line only by the aforementioned mutations, so they can help to understand their impact on the ALS development.
