Control iPSC lines
Control iPSC line – iMA-1L
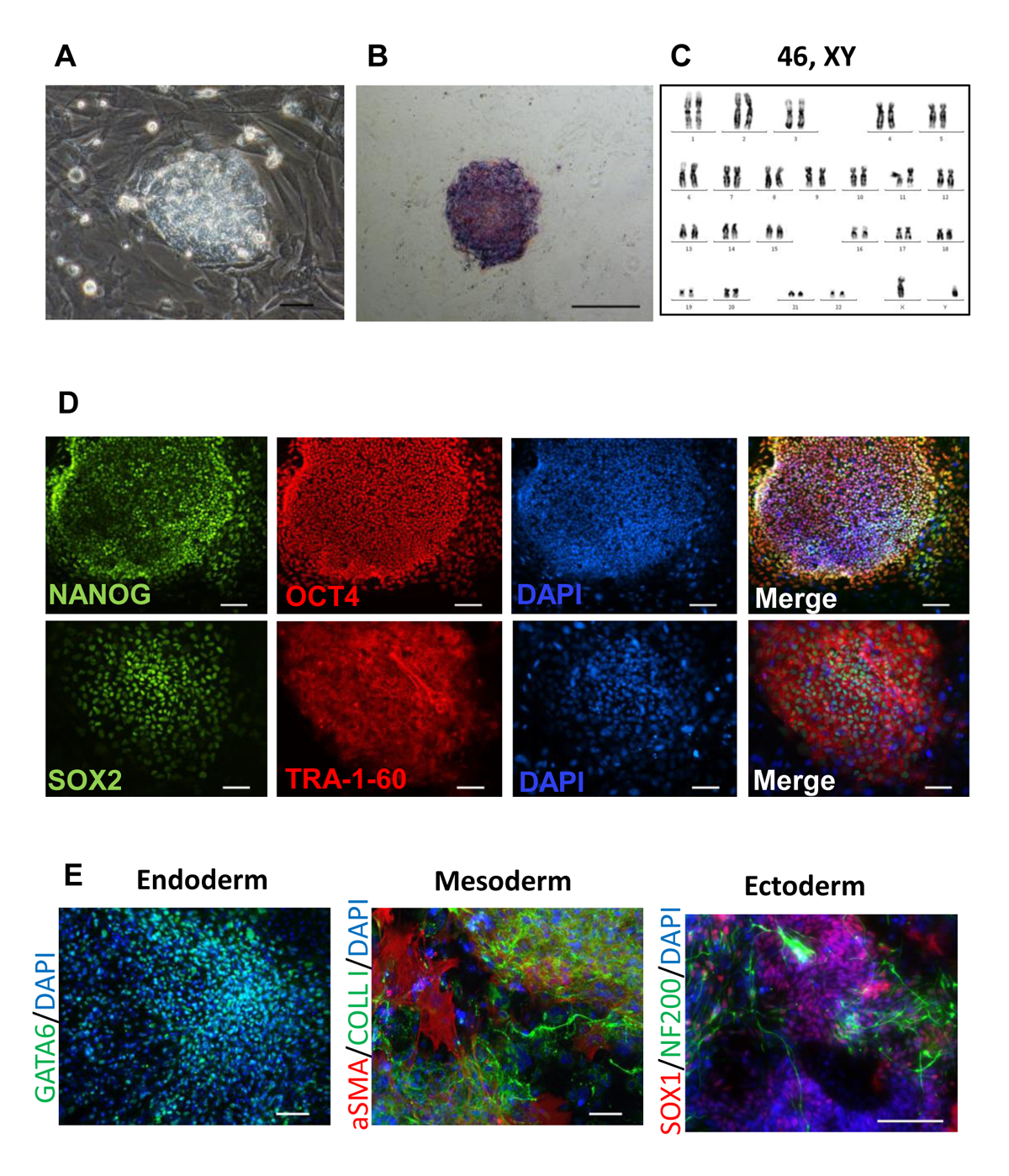
The induced pluripotent stem cell (iPSC) line iMA-1L was generated from human embryonic dermal fibroblasts using episomal vectors expressing pluripotency factors. The episomes were delivered into the cells by nucleofection (NHDF Nucleofector Kit, Lonza). iMA-1L intensively grew in the pluripotent stem cell-like colonies, had dense intercellular contacts, large nuclear-cytoplasmic ratio and expressed endogenous alkaline phosphatase. Immunofluorescent analysis for pluripotency markers showed the expression of OCT4, SOX2, NANOG, and TRA-1-60. iMA-1L has a normal karyotype 46: XY at both early (6) and late (22-24) passages. Immunofluorescent staining showed that iMA-1L can be differentiated into the cells of three germlines. The characteristics of this line was published in Supplementary Information in (Grigor’eva et al., 2020 https://doi.org/10.1007/s10616-020-00406-7).
Control iPSC line – iMA-1L
The induced pluripotent stem cell (iPSC) line iMA-1L was generated from human embryonic dermal fibroblasts using episomal vectors expressing pluripotency factors. The episomes were delivered into the cells by nucleofection (NHDF Nucleofector Kit, Lonza). iMA-1L intensively grew in the pluripotent stem cell-like colonies, had dense intercellular contacts, large nuclear-cytoplasmic ratio and expressed endogenous alkaline phosphatase. Immunofluorescent analysis for pluripotency markers showed the expression of OCT4, SOX2, NANOG, and TRA-1-60. iMA-1L has a normal karyotype 46: XY at both early (6) and late (22-24) passages. Immunofluorescent staining showed that iMA-1L can be differentiated into the cells of three germlines. The characteristics of this line was published in Supplementary Information in (Grigor’eva et al., 2020 https://doi.org/10.1007/s10616-020-00406-7).
Control iPSC lines – ICGi021-A and ICGi022-A
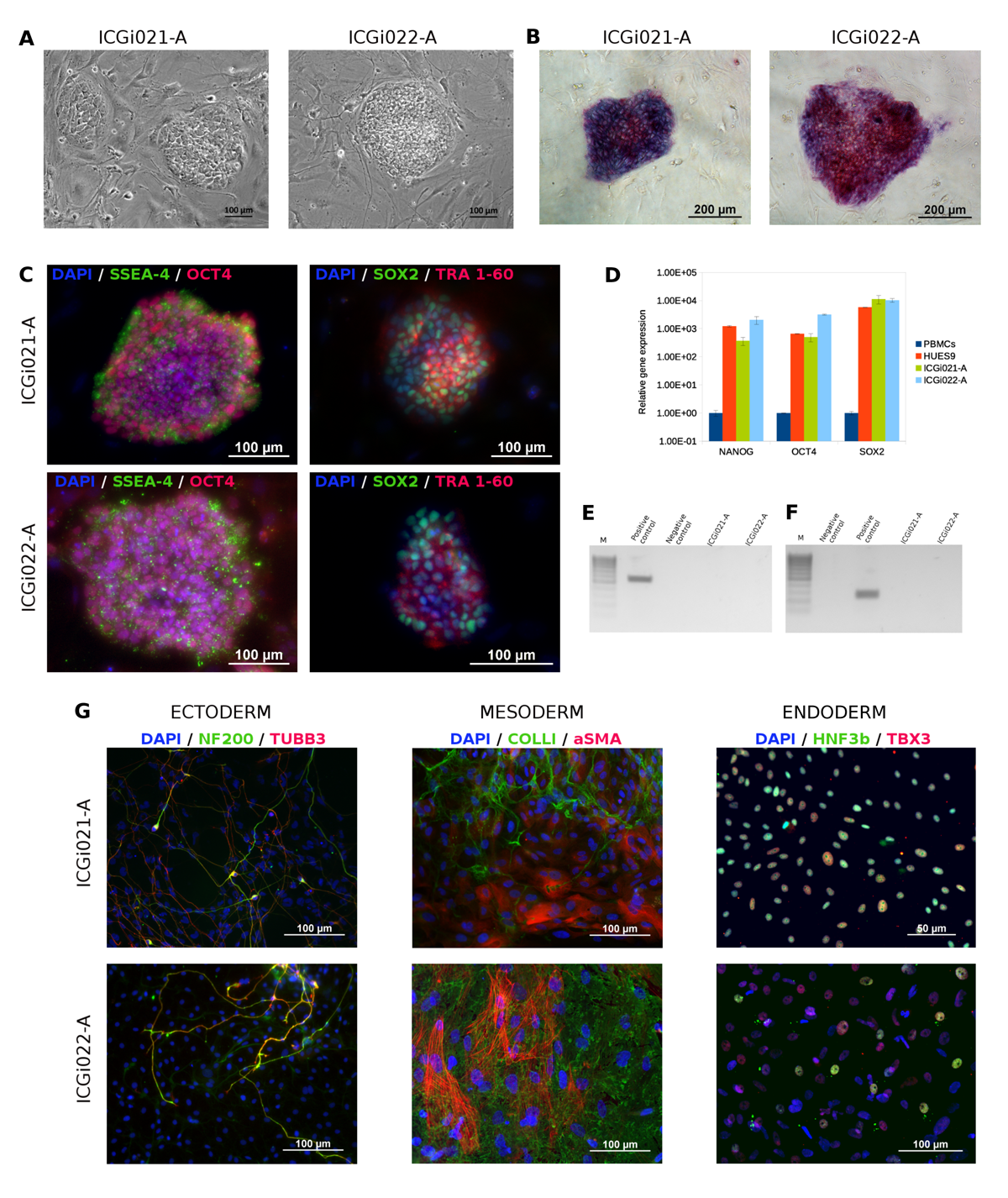
ICGi021-A and ICGi022-A iPSC lines were obtained by reprogramming peripheral blood mononuclear cells from two healthy women of the Siberian population using episomal non-integrating vectors expressing Yamanaka factors. iPSC lines expressed pluripotency markers, have a normal karyotype and demonstrated the ability to differentiate into derivatives of the three germ layers. Clinical exome sequencing data of the original biosamples of the donors are available in the NCBI SRA database (SRR11413028 and SRR11413027). The generated cell lines are useful as “healthy” control in hereditary disease studies, and for basic research of cellular and molecular mechanisms of pluripotency and differentiation.
Control iPSC lines – ICGi021-A and ICGi022-A
ICGi021-A and ICGi022-A iPSC lines were obtained by reprogramming peripheral blood mononuclear cells from two healthy women of the Siberian population using episomal non-integrating vectors expressing Yamanaka factors. iPSC lines expressed pluripotency markers, have a normal karyotype and demonstrated the ability to differentiate into derivatives of the three germ layers. Clinical exome sequencing data of the original biosamples of the donors are available in the NCBI SRA database (SRR11413028 and SRR11413027). The generated cell lines are useful as “healthy” control in hereditary disease studies, and for basic research of cellular and molecular mechanisms of pluripotency and differentiation.
